What are phenotypes in data science? The HDR UK Phenotype Library, explained
26 August 2022
Dan Thayer, a senior data scientist and team lead at the Secure Anonymised Information Linkage (SAIL) databank, introduces the HDR Phenotype Library – explaining what phenotypes are, why they’re so important, and how researchers can contribute to this growing resource.
What is a phenotype?
A phenotype is any observable or measurable trait that an organism has. It could be a physical description – someone’s age, height or weight. It may also be something measured, like blood pressure or glucose levels. Or a phenotype might be a characteristic such as what condition someone has, or the medication they may be taking.
Researchers create these definitions to guide how they use data and interpret its meaning. The phenotypes might look like a list of clinical codes, or they could be an algorithm with a set of rules.
Why are phenotypes needed?
Large health data sources can be extremely valuable for research. Instead of collecting new data, researchers may be able to use an existing big data resource to answer their research question. But how do they find the exact data they need? And in a format they can use it? Often, that’s not straight forward. Phenotypes are a way of solving this problem.
For example, for a study on type 1 diabetes, a researcher might want to look at data from patients diagnosed the condition who are on a certain medication. But perhaps they also want to exclude people with type 2 diabetes, otherwise this could cause an error when interpreting the data.
If there isn’t an easy way to separate these groups within the dataset, this researcher could create a phenotype. That would enable them to find data from people with type 1 diabetes, based on a set of criteria, but exclude those with a type 2 diabetes diagnosis.
How does the HDR UK Phenotype Library help the research community?
The Phenotype Library is a place to share phenotypes between researchers. There is naturally going to be some overlap between studies looking at the same disease area. The HDR UK Phenotype Library makes existing phenotypes visible, so people can discover and reuse them, eliminating the need to repeat work and go over the same ground.
The library also fully captures information about these phenotypes so it’s clear how they’ve been created. For research to be transparent and reproducible, it’s important that there’s a record of exactly what’s been done, and what decisions have been made and why.
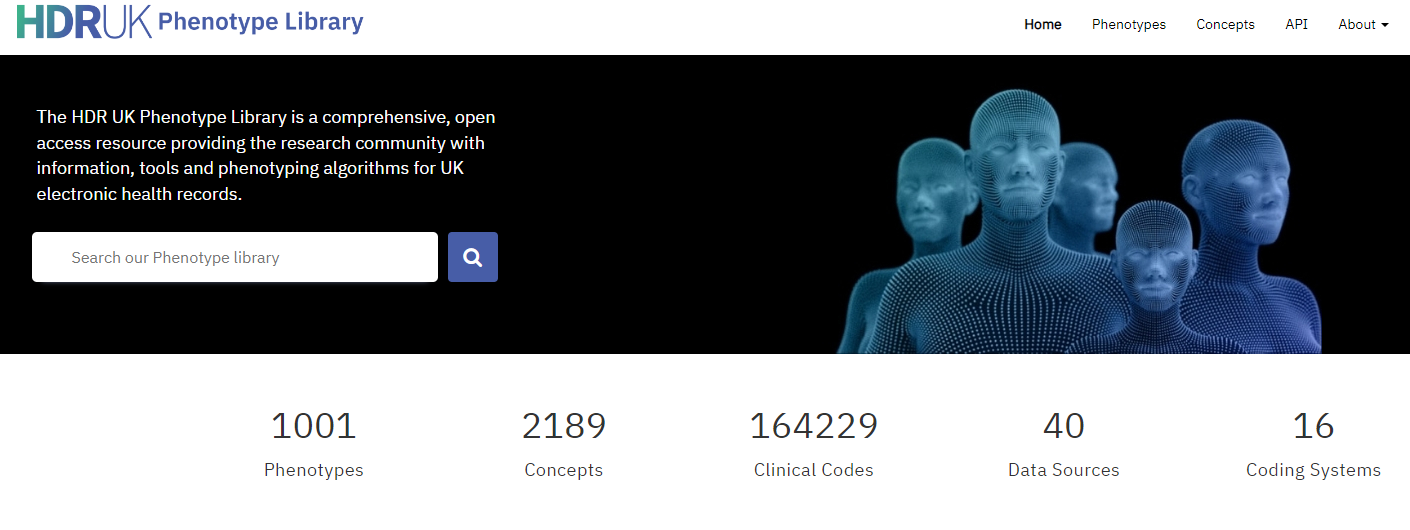
What inspired the development of the Phenotype Library?
As this field of research developed, many of us recognised the need for new tools and methods to support phenotyping and started building solutions. For example, in SAIL we created the Concept Library, a predecessor of the Phenotype Library which met many of the same needs. This HDR UK project was largely about the research community recognising that we need to come together, share what we have been doing, and develop a common approach in this space.
Our aim is for the library to be useful for data scientists and clinicians involved in research. But there may also be aspects of the library that are useful for the public as well. That’s why patient and public involvement has been a core part of the library’s development – asking non-experts to review the site and give feedback on what they understand and what they don’t to make such it’s accessible to all.
How can researchers contribute their work the Phenotype Library?
We want this resource to support as broad an array of research as possible. That’s why we’re keen for more researchers to get involved and contribute their own phenotypes, but also provide feedback.
If researchers are interested in contributing to the library, we encourage them to get in touch and request a user account. If, once registered, they need support to load their content into the library then we are more than happy to help.