How do we collect, store and manage data about people’s SARS-CoV-2 samples?
26 January 2021
Once someone has tested positive for COVID-19 and their sample has been selected for sequencing by The COVID-19 Genomics UK (COG-UK), what happens to their sample data? How do we link individuals’ SARS-CoV-2 sample data with other information, and how do we store and manage all this data?
COG-UK exists to generate and study the SARS-CoV-2 viral genome — its complete set of genetic
information. To learn as much as we can about this new virus, we combine information about its genome with details of when, where and why a sample was taken and about the people whose samples are being tested.
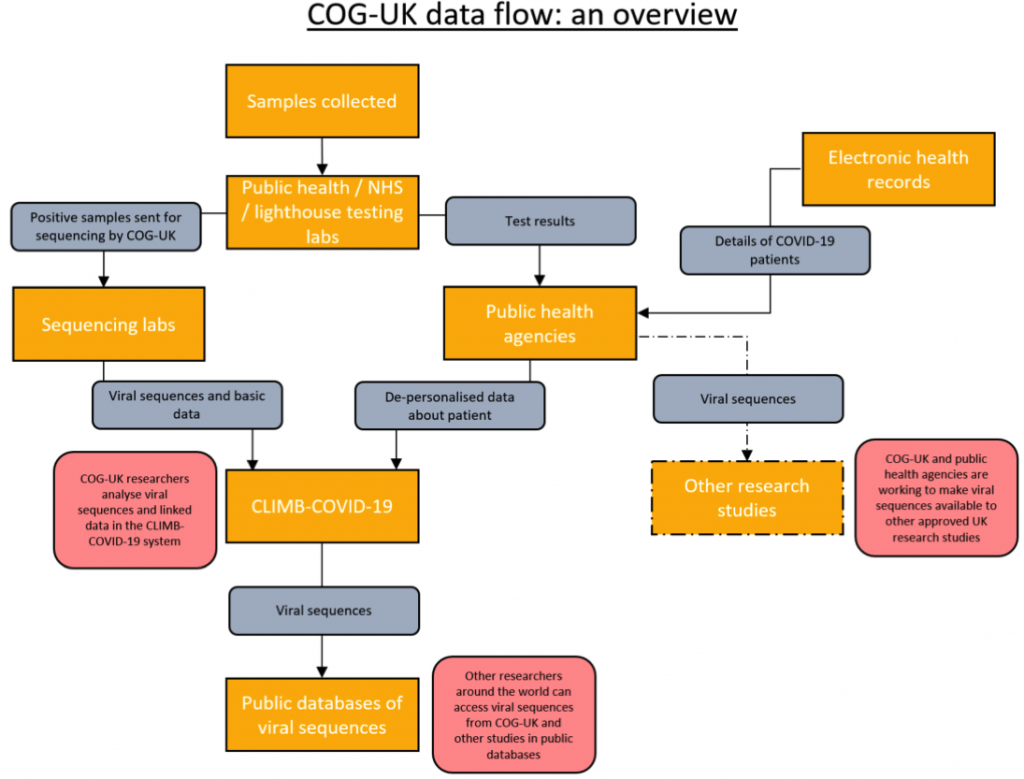
COG-UK has established systems to collect data from labs conducting COVID-19 testing and from the central records held by public health agencies across the UK in ways that keep it secure and protect the confidentiality of people taking tests.
How do we link SARS-CoV-2 sample data with other information?
Each sample submitted to COG-UK is given a new, unique identifier. This consists of a COG-UK site code and a string of letters and numbers — something like COG-4A996. This COG-UK ID is used to link samples, sequences and the associated data together across the consortium.
Labs carrying out tests provide COG-UK with data about when and where the sample was collected as well as basic, non-identifying information about the person who took the test: the first part of their postcode (e.g. SW1A), their age and sex. Labs in hospitals may also be able to say whether the person is a patient, a member of staff or a care home resident.
How do public health agencies use this data?
A record of the COG-UK ID given to each sample is also shared with the public health agency responsible for that part of the UK. Public health agencies use this information to link the viral sequences produced by COG-UK to the other records they hold about people diagnosed with COVID-19. This allows them to use genomics in the public health response.
Public health agencies can also use those links to provide COG-UK with further details about the people whose viral samples have been tested, without COG-UK needing to know their identities. Public health agencies can tell us whether two samples collected at different times or places come from the same person and can give extra, non-identifying information about the sample and person tested.
How do we store this data?
All of this data is combined with the viral sequences in COG-UK’s central database, called CLIMB-COVID-19. The data in this database is de-personalised. This means that anything that directly identifies an individual has been removed. However, we still take great care with how the data is used since particular combinations — like a specific date and place — might allow someone to be identified.
To protect the data and the confidentiality of people diagnosed with COVID-19, the CLIMB-COVID-19 system meets the security standards for processing NHS data. Everyone accessing the system must undergo training in data protection. There are further restrictions on access to the most detailed data and strict rules for what can be included in the results that are removed from the system.
Because public health agencies can connect samples — and so viral sequences — back to individuals they can also provide information about the viral genome for use in other research studies that have permission to access patient data. COG-UK is working with the GenOMICC study, which is sequencing the genomes of volunteers who have been diagnosed with COVID-19. We are also working with HDR UK and The Office for National Statistics to safely make data on viral genetics available to other research studies across the UK.
HDR UK, the nation’s institute for health and biomedical informatics, is contributing to the COG-UK consortium in several ways, including the coordination of national efforts to link COVID-19 genome databases with epidemiological and clinical databases. Through the Health Data Research UK Innovation Gateway, COG-UK has published a dataset that contains over 200,000 SARS-CoV-2 viral genome sequences available as open access to any COVID-19 researchers.
About The COVID-19 Genomics UK (COG-UK)
The current COVID-19 pandemic, caused by the SARS-CoV-2, represents a major threat to health. The COG-UK consortium has been created to deliver large-scale and rapid whole-genome virus sequencing to local NHS centres and the UK government.
Led by Professor Sharon Peacock of the University of Cambridge, COG-UK is made up of an innovative partnership of NHS organisations, the four Public Health Agencies of the UK, 15 academic partners providing sequencing and analysis capacity, and the central sequencing hub of the Wellcome Sanger Institute. A full list of collaborators can be found here. Professor Peacock is also on a part-time secondment to PHE as Director of Science, where she focuses on the development of pathogen sequencing through COG-UK.
COG-UK was established in April 2020 supported by £20 million funding from the COVID-19 rapid-research-response “fighting fund” from Her Majesty’s Treasury (established by Professor Chris Whitty and Sir Patrick Vallance), and administered by the National Institute for Health Research (NIHR), UK Research and Innovation (UKRI), and the Wellcome Sanger Institute. The consortium was also backed by the Department of Health and Social Care’s Testing Innovation Fund on 16 November 2020 to facilitate the genome sequencing capacity needed to meet the increasing number of COVID-19 cases in the UK over the winter period.
Visit COG-UK’s website: www.cogconsortium.uk